Statistical confidence measures for genome maps: application to the validation of genome assemblies
Abstract
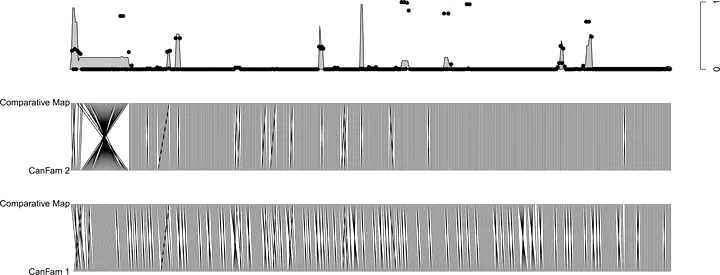
Genome maps are imperative to address the genetic basis of the biology of an organism. While a growing number of genomes are being sequenced providing the ultimate genome maps—this being done at an even faster pace now using new generation sequencers—the process of constructing intermediate maps to build and validate a genome assembly remains an important component for producing complete genome sequences. However, current mapping approach lack statistical confidence measures necessary to identify precisely relevant inconsistencies between a genome map and an assembly. We propose new methods to derive statistical measures of confidence on genome maps using a comparative model for radiation hybrid data. We describe algorithms allowing to (i) sample from a distribution of maps and (ii) exploit this distribution to construct robust maps. We provide an example of application of these methods on a dog dataset that demonstrates the interest of our approach.